
The main command to call our executable program to filter the marker line and export the three result data is as the following: Call an executable program(written by C/C++) to filter the markers that do not meet the specific criterias and output the results as 3 independent files.
The main commands to exclude the SNP/INDEL Markers from scaffold/chloroplast are as the followings:Ĭat bcf_text.txt | grep -v scaffold > bcf_text_exclude.txtĬat bcf_text.txt | grep -v chl > bcf_text_exclude.txtģ. Call some script(mainly written by perl, bash) to exclude some specific markers. The main commands to extract the specific SNP/INDEL markers are as the folowings:īcftools query -H -i'%TYPE="indels"' -f "%CHROM %POS %REF %ALT %DP %QUAL %AF %AC %AN %TYPEīcftools query -H -i'%TYPE="snps"' -f "%CHROM %POS %REF %ALT %DP %QUAL %AF %AC %AN %TYPEĢ. A script to plot single variants is also provided based on ggplot2.
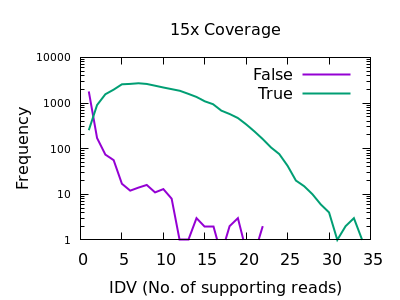
It requires BCFtools 1.14 or newer to run. It can be compiled with BCFtools or downloaded as a set of binary files. Call bcftool to extract the specific SNP/INDEL Markers gtc2vcf is entirely written in C as a BCFtools plugin. In this code, we call vcftools, feed it a vcf file after the -vcf flag, -max-missing 0.5 tells it to filter genotypes called below 50 (across all individuals) the -mac 3 flag. GMEF mainly include the following functional sub-procedures.ġ. In a previous post, I wondered how many pathogenic ClinVar variants were present in ExAC at an allele frequency (AF) greater than 0.01. The first 3 criterias are calculated as a percentages. The specific criterias include: MAF(Minum Allele Frequencey), Heterogenous Genotype Ratio, Unknown Genotype Ratio, and the Allele Length Difference. Calling Variants: Workflow 20 QC Reads and Align Evaluate. from PCR) Both unique and multi-mapped reads are used for calling variants Recalibrates quality scores to take into account sequencing errors 19. CORTEX) Calling Variants: Samtools (cont.) Removes duplicate reads (eg.
#BCFTOOLS FILTER VCF SOFTWARE#
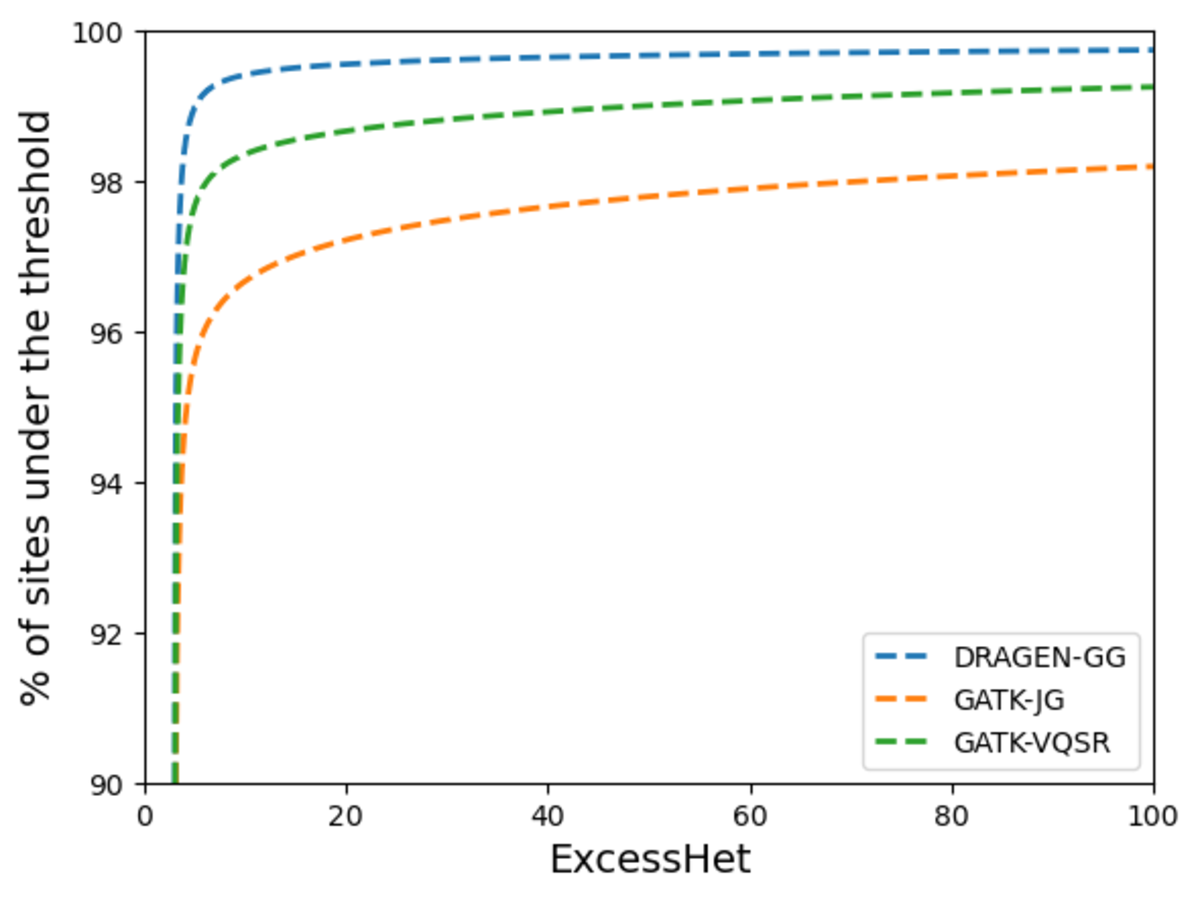
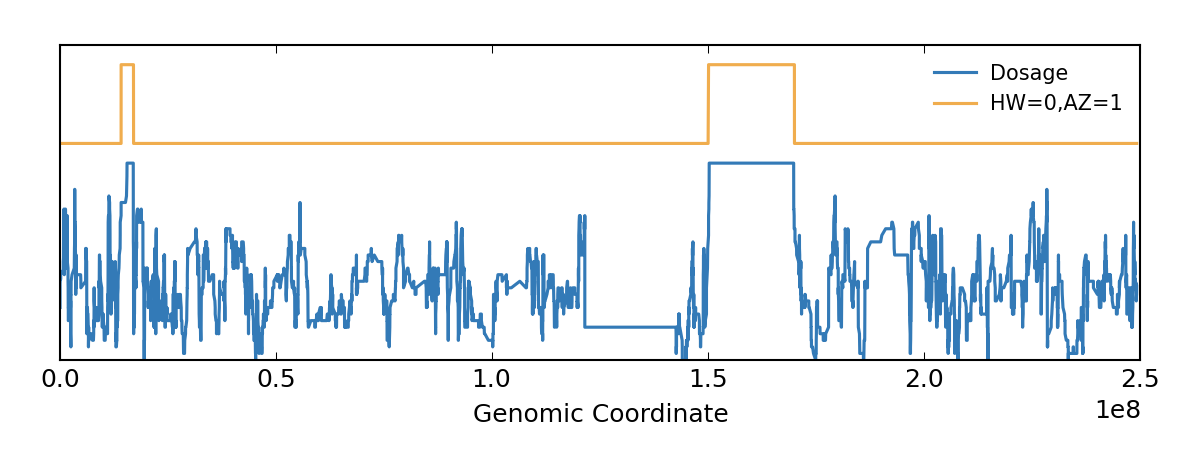
18 reference-free variant calling software are available (eg. ) in bcftools Id like to set low-quality genotypes to missing. Bcftools can be used to filter VCF files. However, we can also run BCFtools to extract more detailed statistics about. Does anyone know if there is a way to do this (set genotypes that dont match the criteria to. The user can specific the marker type as SNP or/and INDEL, whether need to exclude the markers from chloroplast or/and scaffolds, whether the markers meet the specific criterias. Location : variants/ Failed Filters : 0 Passed Filters. The users only need to select the required variant call file, configure the parameters and submit. In GMEF, we have hosted several genetic variant data including the medicago hapmap data. GMEF is a web pipeline, which is aiming to extract and filter the genotype markers from the original genetic variant file, such as. bcftools annotate -x INFO,FORMAT,QUAL,FILTER -o yourfileoutput.vcf yourfile.


 0 kommentar(er)
0 kommentar(er)
